You can run this notebook in a live session or view it on Github.
ROMS Ocean Model Example¶
The Regional Ocean Modeling System (ROMS) is an open source hydrodynamic model that is used for simulating currents and water properties in coastal and estuarine regions. ROMS is one of a few standard ocean models, and it has an active user community.
ROMS uses a regular C-Grid in the horizontal, similar to other structured grid ocean and atmospheric models, and a stretched vertical coordinate (see the ROMS documentation for more details). Both of these require special treatment when using xarray to analyze ROMS ocean model output. This example notebook shows how to create a lazily evaluated vertical coordinate, and make some basic plots. The xgcm package is required to do
analysis that is aware of the horizontal C-Grid.
[1]:
import numpy as np
import cartopy.crs as ccrs
import cartopy.feature as cfeature
import matplotlib.pyplot as plt
%matplotlib inline
import xarray as xr
Load a sample ROMS file. This is a subset of a full model available at
http://barataria.tamu.edu/thredds/catalog.html?dataset=txla_hindcast_agg
The subsetting was done using the following command on one of the output files:
#open dataset
ds = xr.open_dataset('/d2/shared/TXLA_ROMS/output_20yr_obc/2001/ocean_his_0015.nc')
# Turn on chunking to activate dask and parallelize read/write.
ds = ds.chunk({'ocean_time': 1})
# Pick out some of the variables that will be included as coordinates
ds = ds.set_coords(['Cs_r', 'Cs_w', 'hc', 'h', 'Vtransform'])
# Select a a subset of variables. Salt will be visualized, zeta is used to
# calculate the vertical coordinate
variables = ['salt', 'zeta']
ds[variables].isel(ocean_time=slice(47, None, 7*24),
xi_rho=slice(300, None)).to_netcdf('ROMS_example.nc', mode='w')
So, the ROMS_example.nc file contains a subset of the grid, one 3D variable, and two time steps.
Load in ROMS dataset as an xarray object¶
[2]:
# load in the file
ds = xr.tutorial.open_dataset('ROMS_example.nc', chunks={'ocean_time': 1})
# This is a way to turn on chunking and lazy evaluation. Opening with mfdataset, or
# setting the chunking in the open_dataset would also achive this.
ds
[2]:
- eta_rho: 191
- ocean_time: 2
- s_rho: 30
- xi_rho: 371
- Cs_r(s_rho)float64dask.array<chunksize=(30,), meta=np.ndarray>
- long_name :
- S-coordinate stretching curves at RHO-points
- valid_min :
- -1.0
- valid_max :
- 0.0
- field :
- Cs_r, scalar
Array Chunk Bytes 240 B 240 B Shape (30,) (30,) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - lon_rho(eta_rho, xi_rho)float64dask.array<chunksize=(191, 371), meta=np.ndarray>
- long_name :
- longitude of RHO-points
- units :
- degree_east
- standard_name :
- longitude
- field :
- lon_rho, scalar
Array Chunk Bytes 566.89 kB 566.89 kB Shape (191, 371) (191, 371) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - hc()float64...
- long_name :
- S-coordinate parameter, critical depth
- units :
- meter
array(20.)
- h(eta_rho, xi_rho)float64dask.array<chunksize=(191, 371), meta=np.ndarray>
- long_name :
- bathymetry at RHO-points
- units :
- meter
- field :
- bath, scalar
Array Chunk Bytes 566.89 kB 566.89 kB Shape (191, 371) (191, 371) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - lat_rho(eta_rho, xi_rho)float64dask.array<chunksize=(191, 371), meta=np.ndarray>
- long_name :
- latitude of RHO-points
- units :
- degree_north
- standard_name :
- latitude
- field :
- lat_rho, scalar
Array Chunk Bytes 566.89 kB 566.89 kB Shape (191, 371) (191, 371) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - Vtransform()int32...
- long_name :
- vertical terrain-following transformation equation
array(2, dtype=int32)
- ocean_time(ocean_time)datetime64[ns]2001-08-01 2001-08-08
- long_name :
- time since initialization
- field :
- time, scalar, series
array(['2001-08-01T00:00:00.000000000', '2001-08-08T00:00:00.000000000'], dtype='datetime64[ns]') - s_rho(s_rho)float64-0.9833 -0.95 ... -0.05 -0.01667
- long_name :
- S-coordinate at RHO-points
- valid_min :
- -1.0
- valid_max :
- 0.0
- positive :
- up
- standard_name :
- ocean_s_coordinate_g2
- formula_terms :
- s: s_rho C: Cs_r eta: zeta depth: h depth_c: hc
- field :
- s_rho, scalar
array([-0.983333, -0.95 , -0.916667, -0.883333, -0.85 , -0.816667, -0.783333, -0.75 , -0.716667, -0.683333, -0.65 , -0.616667, -0.583333, -0.55 , -0.516667, -0.483333, -0.45 , -0.416667, -0.383333, -0.35 , -0.316667, -0.283333, -0.25 , -0.216667, -0.183333, -0.15 , -0.116667, -0.083333, -0.05 , -0.016667])
- salt(ocean_time, s_rho, eta_rho, xi_rho)float32dask.array<chunksize=(1, 30, 191, 371), meta=np.ndarray>
- long_name :
- salinity
- time :
- ocean_time
- field :
- salinity, scalar, series
Array Chunk Bytes 17.01 MB 8.50 MB Shape (2, 30, 191, 371) (1, 30, 191, 371) Count 3 Tasks 2 Chunks Type float32 numpy.ndarray - zeta(ocean_time, eta_rho, xi_rho)float32dask.array<chunksize=(1, 191, 371), meta=np.ndarray>
- long_name :
- free-surface
- units :
- meter
- time :
- ocean_time
- field :
- free-surface, scalar, series
Array Chunk Bytes 566.89 kB 283.44 kB Shape (2, 191, 371) (1, 191, 371) Count 3 Tasks 2 Chunks Type float32 numpy.ndarray
- file :
- ../output_20yr_obc/2001/ocean_his_0015.nc
- format :
- netCDF-4/HDF5 file
- Conventions :
- CF-1.4
- type :
- ROMS/TOMS history file
- title :
- TXLA ROMS hindcast run with dyes and oxygen
- rst_file :
- ../output_20yr_obc/2001/ocean_rst.nc
- his_base :
- ../output_20yr_obc/2001/ocean_his
- avg_base :
- ../output_20yr_obc/2001/ocean_avg
- dia_base :
- ../output_20yr_obc/2001/ocean_dia
- sta_file :
- ocean_sta.nc
- grd_file :
- ../inputs/grd/txla2_grd_v4_test_lcut_hglo_wtype.nc
- ini_file :
- ../output_20yr_obc/2000/ocean_rst.nc
- frc_file_01 :
- ../inputs/2001/txla_bulk_ERAI_2001.nc
- frc_file_02 :
- ../inputs/2001/txla_flx_ICOADS_AVHRR_SST_2001.nc, ../inputs/2002/txla_flx_ICOADS_AVHRR_SST_2002.nc
- bry_file_01 :
- ../inputs/2001/txla2_bry_2001_glo_ReAna_v4_o2woa.nc, ../inputs/2002/txla2_bry_2002_glo_ReAna_v4_o2woa.nc
- clm_file_01 :
- ../inputs/2001/txla2_clm_2001_01_glo_ReAna_v4_o2woa.nc, ../inputs/2001/txla2_clm_2001_02_glo_ReAna_v4_o2woa.nc, ../inputs/2001/txla2_clm_2001_03_glo_ReAna_v4_o2woa.nc, ../inputs/2001/txla2_clm_2001_04_glo_ReAna_v4_o2woa.nc, ../inputs/2001/txla2_clm_2001_05_glo_ReAna_v4_o2woa.nc, ../inputs/2001/txla2_clm_2001_06_glo_ReAna_v4_o2woa.nc, ../inputs/2001/txla2_clm_2001_07_glo_ReAna_v4_o2woa.nc, ../inputs/2001/txla2_clm_2001_08_glo_ReAna_v4_o2woa.nc, ../inputs/2001/txla2_clm_2001_09_glo_ReAna_v4_o2woa.nc, ../inputs/2001/txla2_clm_2001_10_glo_ReAna_v4_o2woa.nc, ../inputs/2001/txla2_clm_2001_11_glo_ReAna_v4_o2woa.nc, ../inputs/2001/txla2_clm_2001_12_glo_ReAna_v4_o2woa.nc, ../inputs/2002/txla2_clm_2002_01_glo_ReAna_v4_o2woa.nc
- script_file :
- spos_file :
- /scratch/user/d.kobashi/inputs/stations.in
- NLM_LBC :
- EDGE: WEST SOUTH EAST NORTH zeta: Che Che Che Clo ubar: Shc Shc Shc Clo vbar: Shc Shc Shc Clo u: Rad Rad Rad Clo v: Rad Rad Rad Clo temp: Rad Rad Rad Clo salt: Rad Rad Rad Clo dye_01: Gra Gra Gra Clo dye_02: Rad Rad Rad Clo dye_03: Rad Rad Rad Clo dye_04: Rad Rad Rad Clo tke: Gra Gra Gra Clo
- svn_url :
- https:://myroms.org/svn/src
- svn_rev :
- code_dir :
- /scratch/user/d.kobashi/source_code/COAWST/COAWST.r960-dev
- header_dir :
- /home/d.kobashi/TXLA_ROMS_reana/work_20yr_obc
- header_file :
- txla2.h
- os :
- Linux
- cpu :
- x86_64
- compiler_system :
- ifort
- compiler_command :
- /software/easybuild/software/impi/5.0.1.035-iccifort-2015.0.090/bin64/mpiifort
- compiler_flags :
- -heap-arrays -fp-model fast -mt_mpi -ip -O3 -msse2 -free
- tiling :
- 010x012
- history :
- Tue Jul 24 11:04:43 2018: /opt/nco/ncks -D 4 -t 8 /copano/d1/shared/TXLA_ROMS/output_20yr_obc/2001/ocean_his_0015.nc --cnk_dmn ocean_time,4 --cnk_dmn eta_rho,8 --cnk_dmn eta_u,8 --cnk_dmn eta_v,8 --cnk_dmn eta_psi,8 --cnk_dmn xi_rho,16 --cnk_dmn xi_u,16 --cnk_dmn xi_v,16 --cnk_dmn xi_psi,16 --cnk_dmn s_rho,2 --cnk_dmn s_w,2 --output /copano/d2/shared/TXLA_ROMS/output_20yr_obc/2001/ocean_his_0015.nc ROMS/TOMS, Version 3.7, Monday - July 18, 2016 - 10:38:26 PM
- ana_file :
- /home/d.kobashi/TXLA_ROMS_reana/Functionals/ana_btflux.h, /home/d.kobashi/TXLA_ROMS_reana/Functionals/ana_sponge.h, /home/d.kobashi/TXLA_ROMS_reana/Functionals/ana_nudgcoef.h, /home/d.kobashi/TXLA_ROMS_reana/Functionals/ana_stflux.h
- CPP_options :
- TXLA2, ANA_BPFLUX, ANA_BSFLUX, ANA_BTFLUX, ANA_NUDGCOEF, ANA_SPFLUX, ANA_SPONGE, ASSUMED_SHAPE, AVERAGES, BULK_FLUXES, CURVGRID, DEFLATE, DIAGNOSTICS_TS, DIAGNOSTICS_UV, DIFF_GRID, DJ_GRADPS, DOUBLE_PRECISION, EMINUSP, GLS_MIXING, HDF5, KANTHA_CLAYSON, LONGWAVE, MASKING, MIX_GEO_TS, MIX_S_UV, MPI, NONLINEAR, NONLIN_EOS, N2S2_HORAVG, POWER_LAW, PROFILE, QCORRECTION, K_GSCHEME, RADIATION_2D, !RST_SINGLE, SALINITY, SOLAR_SOURCE, SOLVE3D, SPLINES, SPHERICAL, STATIONS, T_PASSIVE, TS_MPDATA, TS_DIF2, UV_ADV, UV_COR, UV_U3HADVECTION, UV_C4VADVECTION, UV_LOGDRAG, UV_VIS2, VAR_RHO_2D, VISC_GRID, WTYPE_GRID
- NCO :
- netCDF Operators version 4.7.6-alpha04 (Homepage = http://nco.sf.net, Code = http://github.com/nco/nco)
Add a lazilly calculated vertical coordinates¶
Write equations to calculate the vertical coordinate. These will be only evaluated when data is requested. Information about the ROMS vertical coordinate can be found (here)[https://www.myroms.org/wiki/Vertical_S-coordinate]
In short, for Vtransform==2 as used in this example,
\(Z_0 = (h_c \, S + h \,C) / (h_c + h)\)
\(z = Z_0 (\zeta + h) + \zeta\)
where the variables are defined as in the link above.
[3]:
if ds.Vtransform == 1:
Zo_rho = ds.hc * (ds.s_rho - ds.Cs_r) + ds.Cs_r * ds.h
z_rho = Zo_rho + ds.zeta * (1 + Zo_rho/ds.h)
elif ds.Vtransform == 2:
Zo_rho = (ds.hc * ds.s_rho + ds.Cs_r * ds.h) / (ds.hc + ds.h)
z_rho = ds.zeta + (ds.zeta + ds.h) * Zo_rho
ds.coords['z_rho'] = z_rho.transpose() # needing transpose seems to be an xarray bug
ds.salt
<ipython-input-3-8742ef454e3d>:8: FutureWarning: This DataArray contains multi-dimensional coordinates. In the future, these coordinates will be transposed as well unless you specify transpose_coords=False.
ds.coords['z_rho'] = z_rho.transpose() # needing transpose seems to be an xarray bug
[3]:
- ocean_time: 2
- s_rho: 30
- eta_rho: 191
- xi_rho: 371
- dask.array<chunksize=(1, 30, 191, 371), meta=np.ndarray>
Array Chunk Bytes 17.01 MB 8.50 MB Shape (2, 30, 191, 371) (1, 30, 191, 371) Count 3 Tasks 2 Chunks Type float32 numpy.ndarray - Cs_r(s_rho)float64dask.array<chunksize=(30,), meta=np.ndarray>
- long_name :
- S-coordinate stretching curves at RHO-points
- valid_min :
- -1.0
- valid_max :
- 0.0
- field :
- Cs_r, scalar
Array Chunk Bytes 240 B 240 B Shape (30,) (30,) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - lon_rho(eta_rho, xi_rho)float64dask.array<chunksize=(191, 371), meta=np.ndarray>
- long_name :
- longitude of RHO-points
- units :
- degree_east
- standard_name :
- longitude
- field :
- lon_rho, scalar
Array Chunk Bytes 566.89 kB 566.89 kB Shape (191, 371) (191, 371) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - hc()float6420.0
- long_name :
- S-coordinate parameter, critical depth
- units :
- meter
array(20.)
- h(eta_rho, xi_rho)float64dask.array<chunksize=(191, 371), meta=np.ndarray>
- long_name :
- bathymetry at RHO-points
- units :
- meter
- field :
- bath, scalar
Array Chunk Bytes 566.89 kB 566.89 kB Shape (191, 371) (191, 371) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - lat_rho(eta_rho, xi_rho)float64dask.array<chunksize=(191, 371), meta=np.ndarray>
- long_name :
- latitude of RHO-points
- units :
- degree_north
- standard_name :
- latitude
- field :
- lat_rho, scalar
Array Chunk Bytes 566.89 kB 566.89 kB Shape (191, 371) (191, 371) Count 2 Tasks 1 Chunks Type float64 numpy.ndarray - Vtransform()int322
- long_name :
- vertical terrain-following transformation equation
array(2, dtype=int32)
- ocean_time(ocean_time)datetime64[ns]2001-08-01 2001-08-08
- long_name :
- time since initialization
- field :
- time, scalar, series
array(['2001-08-01T00:00:00.000000000', '2001-08-08T00:00:00.000000000'], dtype='datetime64[ns]') - s_rho(s_rho)float64-0.9833 -0.95 ... -0.05 -0.01667
- long_name :
- S-coordinate at RHO-points
- valid_min :
- -1.0
- valid_max :
- 0.0
- positive :
- up
- standard_name :
- ocean_s_coordinate_g2
- formula_terms :
- s: s_rho C: Cs_r eta: zeta depth: h depth_c: hc
- field :
- s_rho, scalar
array([-0.983333, -0.95 , -0.916667, -0.883333, -0.85 , -0.816667, -0.783333, -0.75 , -0.716667, -0.683333, -0.65 , -0.616667, -0.583333, -0.55 , -0.516667, -0.483333, -0.45 , -0.416667, -0.383333, -0.35 , -0.316667, -0.283333, -0.25 , -0.216667, -0.183333, -0.15 , -0.116667, -0.083333, -0.05 , -0.016667]) - z_rho(s_rho, xi_rho, eta_rho, ocean_time)float64dask.array<chunksize=(30, 371, 191, 1), meta=np.ndarray>
Array Chunk Bytes 34.01 MB 17.01 MB Shape (30, 371, 191, 2) (30, 371, 191, 1) Count 35 Tasks 2 Chunks Type float64 numpy.ndarray
- long_name :
- salinity
- time :
- ocean_time
- field :
- salinity, scalar, series
A naive vertical slice¶
Create a slice using the s-coordinate as the vertical dimension is typically not very informative.
[4]:
ds.salt.isel(xi_rho=50, ocean_time=0).plot()
[4]:
<matplotlib.collections.QuadMesh at 0x7f1ce22a2a30>
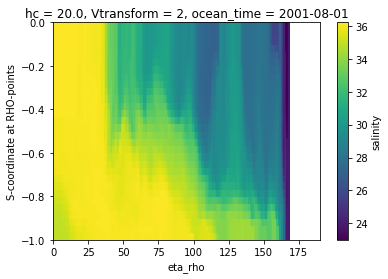
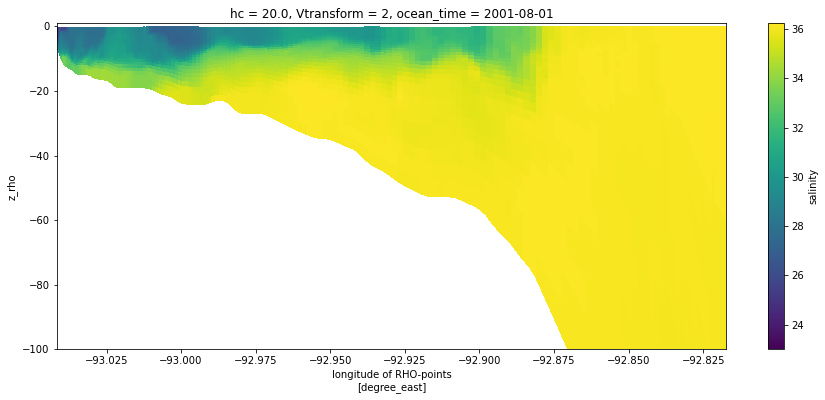
We can feed coordinate information to the plot method to give a more informative cross-section that uses the depths. Note that we did not need to slice the depth or longitude information separately, this was done automatically as the variable was sliced.
[5]:
section = ds.salt.isel(xi_rho=50, eta_rho=slice(0, 167), ocean_time=0)
section.plot(x='lon_rho', y='z_rho', figsize=(15, 6), clim=(25, 35))
plt.ylim([-100, 1]);
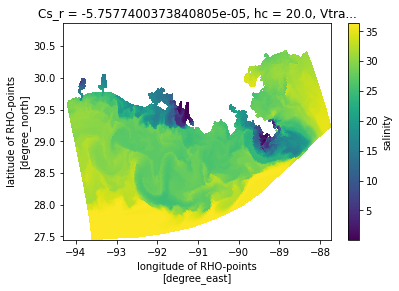
A plan view¶
Now make a naive plan view, without any projection information, just using lon/lat as x/y. This looks OK, but will appear compressed because lon and lat do not have an aspect constrained by the projection.
[6]:
ds.salt.isel(s_rho=-1, ocean_time=0).plot(x='lon_rho', y='lat_rho')
[6]:
<matplotlib.collections.QuadMesh at 0x7f1ce20a4fd0>
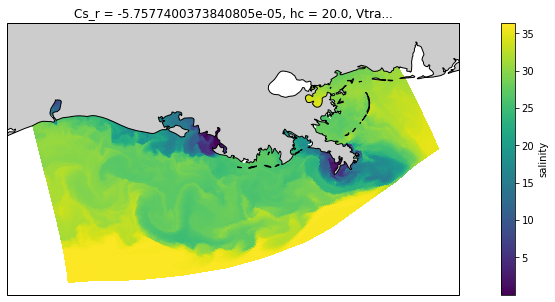
And let’s use a projection to make it nicer, and add a coast.
[7]:
proj = ccrs.LambertConformal(central_longitude=-92, central_latitude=29)
fig = plt.figure(figsize=(15, 5))
ax = plt.axes(projection=proj)
ds.salt.isel(s_rho=-1, ocean_time=0).plot(x='lon_rho', y='lat_rho',
transform=ccrs.PlateCarree())
coast_10m = cfeature.NaturalEarthFeature('physical', 'land', '10m',
edgecolor='k', facecolor='0.8')
ax.add_feature(coast_10m)
[7]:
<cartopy.mpl.feature_artist.FeatureArtist at 0x7f1ce21bc8e0>
/home/docs/checkouts/readthedocs.org/user_builds/xray/conda/v0.15.1/lib/python3.8/site-packages/cartopy/io/__init__.py:260: DownloadWarning: Downloading: http://naciscdn.org/naturalearth/10m/physical/ne_10m_land.zip
warnings.warn('Downloading: {}'.format(url), DownloadWarning)
[ ]: